Nayan P. Agarwal, Michael Matthies, Fatih F. N. Gür, Kensuke Osada, Thorsten L. Schmidt
Angew. Chem. Int. Ed. 2017, DOI:10.1002/anie.201608873

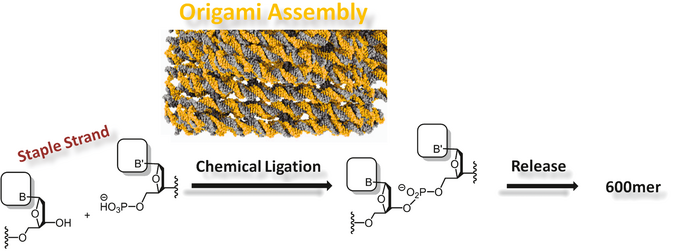
DNA nanotechnology enables the synthesis of nanometer-sized objects that can be site-specifically functionalized with a large variety of materials. For these reasons, DNA-based devices such as DNA origami are being considered for applications in molecular biology and nanomedicine. However, many DNA structures need a higher ionic strength than that of common cell culture buffers or bodily fluids to maintain their integrity and can be degraded quickly by nucleases. To overcome these deficiencies, we coated several different DNA origami structures with a cationic poly(ethylene glycol)–polylysine block copolymer, which electrostatically covered the DNA nanostructures to form DNA origami polyplex micelles (DOPMs). This straightforward, cost-effective, and robust route to protect DNA-based structures could therefore enable applications in biology and nanomedicine where unprotected DNA origami would be degraded.