Paper Highlights
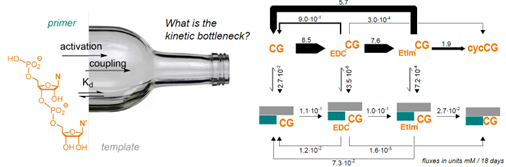
- A quantitative model of enzyme-free copying of RNA with dimers
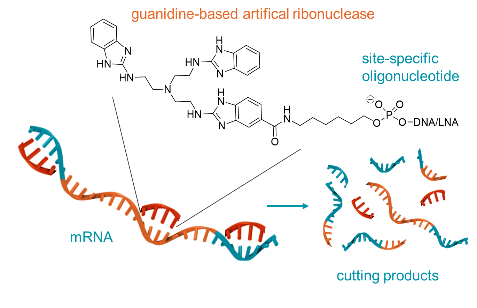
- Quality Control of mRNA Vaccines by Synthetic Ribonucleases: Analysis of the Poly-A-Tail
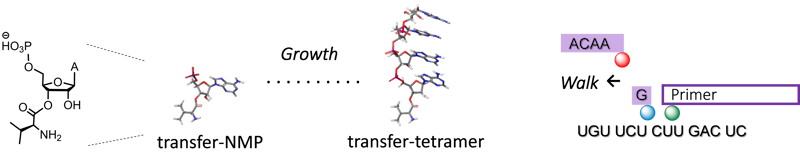
- Ribosome-free translation up to pentapeptides via template walkon RNA sequences
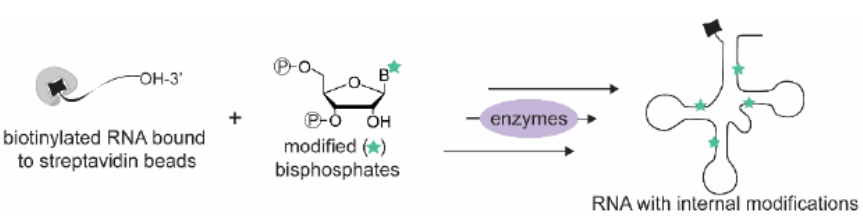
- Solid-Phase-Supported Chemoenzymatic Synthesis of a Light-Activatable tRNA Derivative
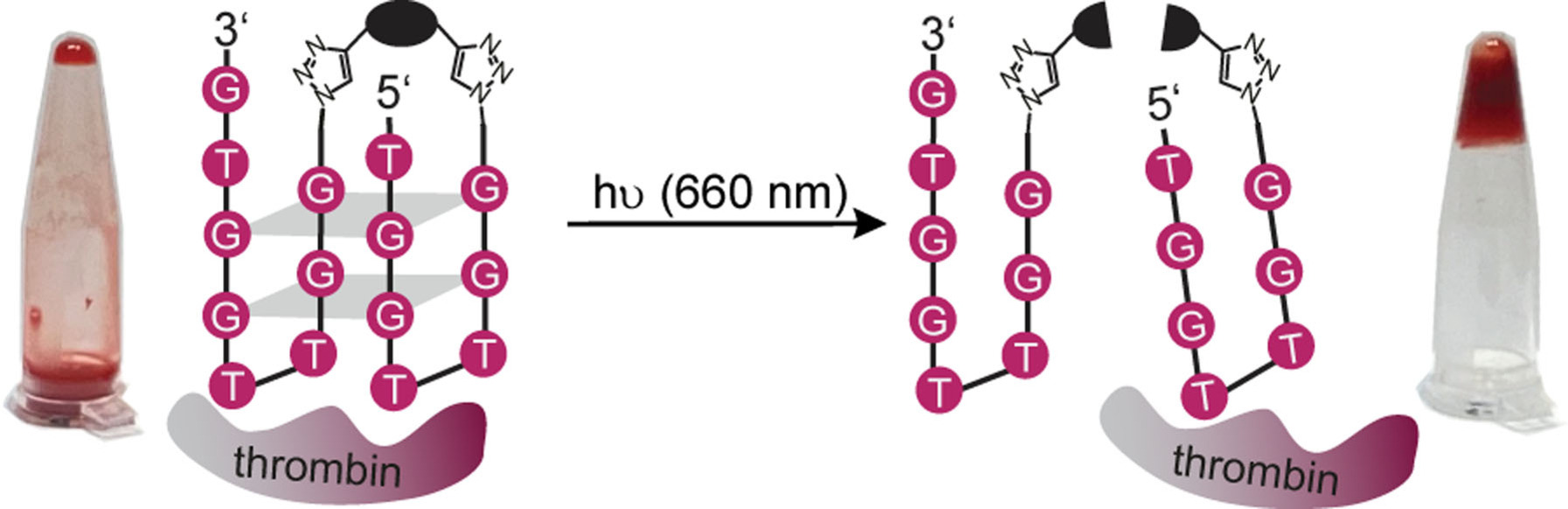
- Controlling Coagulation in Blood with Red Light